Networks
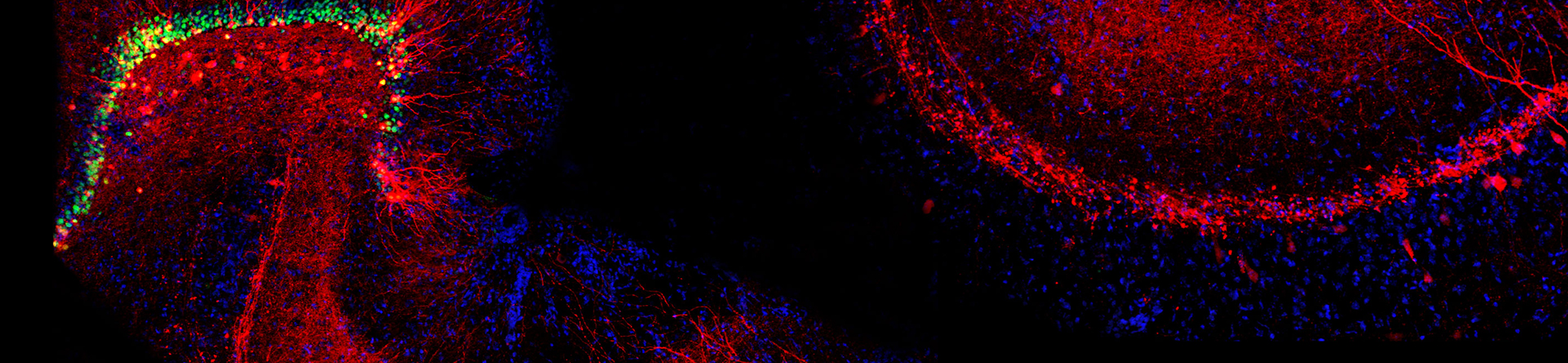
 Over the past decade, a host of new approaches have provided my lab with powerful opportunities to identify novel transporter-associated proteins as well as genes whose activity is regulated by, or can regulate, synaptic signaling. Using the powerful genetic model system Caenorhabitis elegans, and taking advantage of behaviors that emerge with loss of the dopamine transporter, we have implemented unbiased genetic screens that have led to the identification of genes required for the precise regulation of dopamine signaling. The genes identified in these screens have mammalian counterparts and so their identification opens up new opportunities to understand the molecular control of dopamine signaling as well as potential insights into risk for brain disorders associated with compromised dopamine signaling such as ADHD, schizophrenia and addiction. A second unbiased approach involves the use of proteomic methods to identify both transporter associated proteins and the impact of transporter mutations more broadly on the brain proteome. These studies take advantage of capabilities at Vanderbilt to utilize high dimensional mass spectrometry-based approaches pursued on biological extracts as well as on brain sections, where our studies afford evaluation of the impact of transporter mutations with inspection of key brain regions involved simultaneously. Lastly, whole transcriptome sequencing approaches, we pursue the elucidation of gene expression networks that arise from either natural or engineered neurotransmitter transporter sequence variation, as well as drug manipulations. These studies offer high-content data streams that can be mined for unsuspected gene expression relationships that may be key in advancing a broader approach to the assignment of networks of risk genes for brain disorders as well as the development of new medications.
Over the past decade, a host of new approaches have provided my lab with powerful opportunities to identify novel transporter-associated proteins as well as genes whose activity is regulated by, or can regulate, synaptic signaling. Using the powerful genetic model system Caenorhabitis elegans, and taking advantage of behaviors that emerge with loss of the dopamine transporter, we have implemented unbiased genetic screens that have led to the identification of genes required for the precise regulation of dopamine signaling. The genes identified in these screens have mammalian counterparts and so their identification opens up new opportunities to understand the molecular control of dopamine signaling as well as potential insights into risk for brain disorders associated with compromised dopamine signaling such as ADHD, schizophrenia and addiction. A second unbiased approach involves the use of proteomic methods to identify both transporter associated proteins and the impact of transporter mutations more broadly on the brain proteome. These studies take advantage of capabilities at Vanderbilt to utilize high dimensional mass spectrometry-based approaches pursued on biological extracts as well as on brain sections, where our studies afford evaluation of the impact of transporter mutations with inspection of key brain regions involved simultaneously. Lastly, whole transcriptome sequencing approaches, we pursue the elucidation of gene expression networks that arise from either natural or engineered neurotransmitter transporter sequence variation, as well as drug manipulations. These studies offer high-content data streams that can be mined for unsuspected gene expression relationships that may be key in advancing a broader approach to the assignment of networks of risk genes for brain disorders as well as the development of new medications.